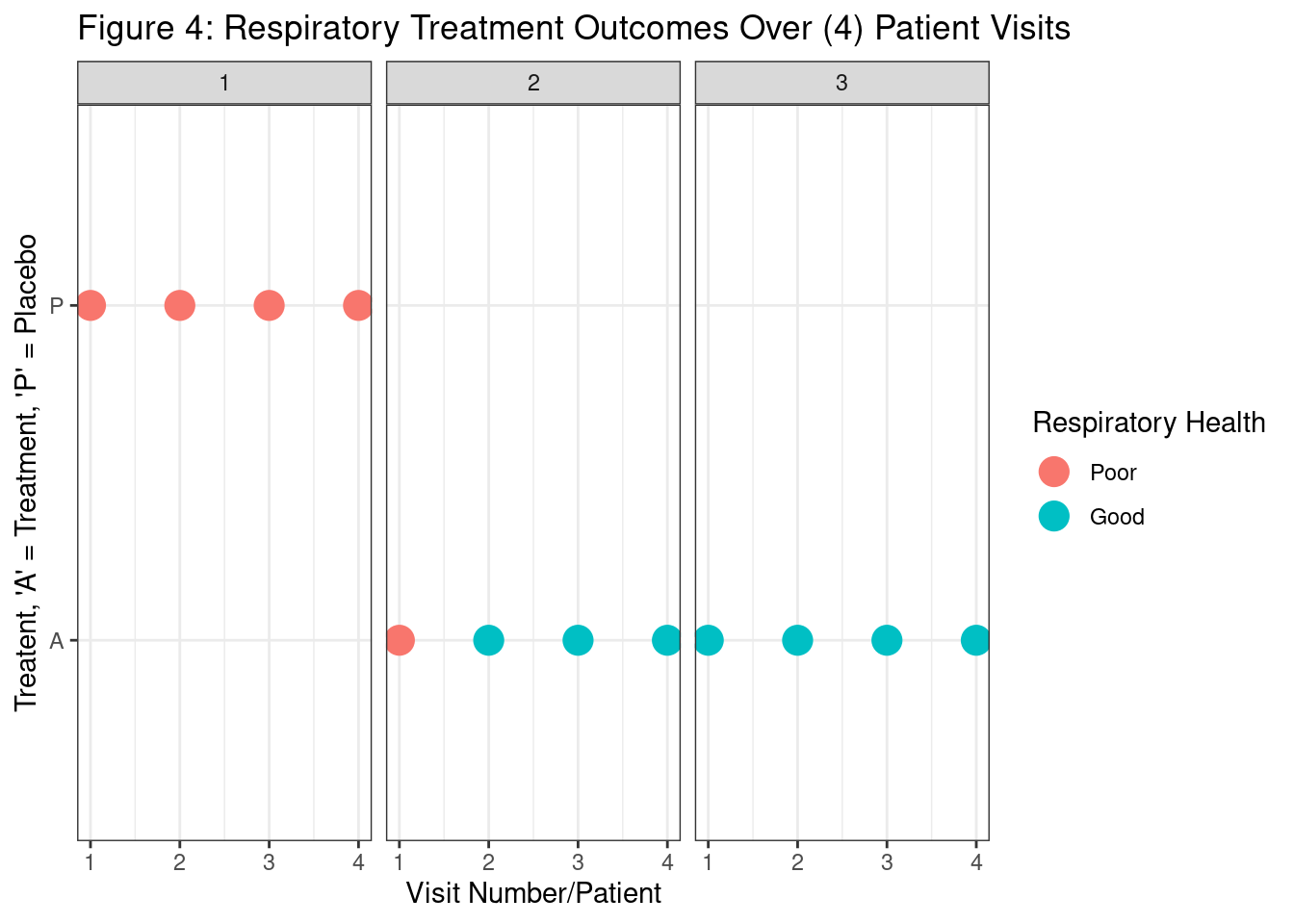
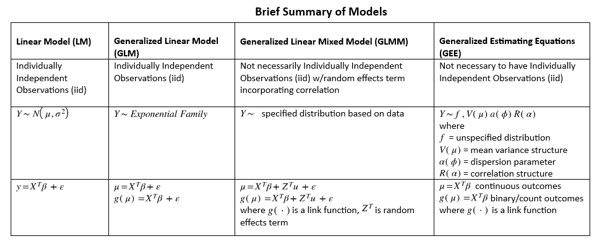
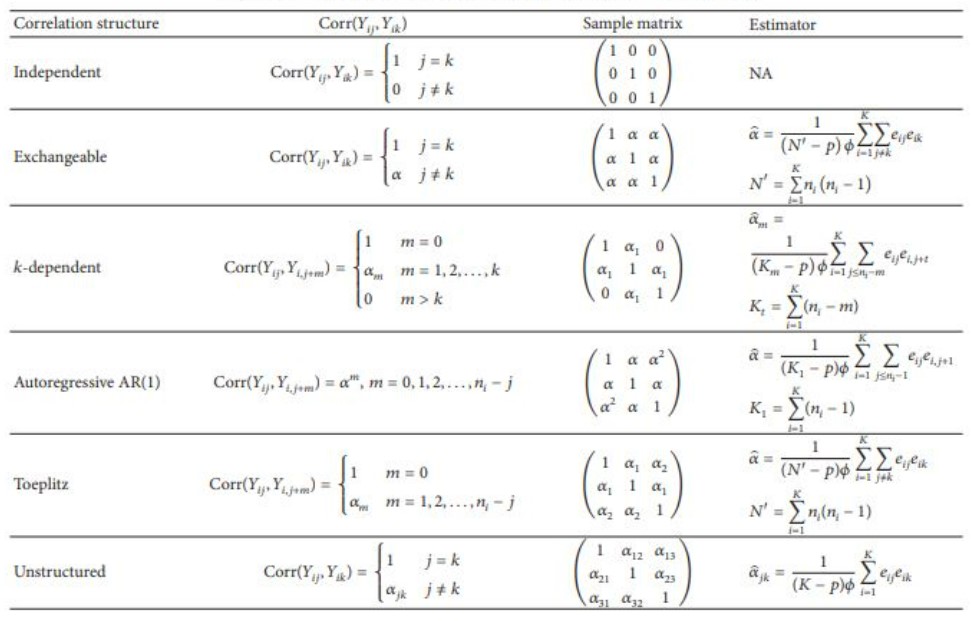
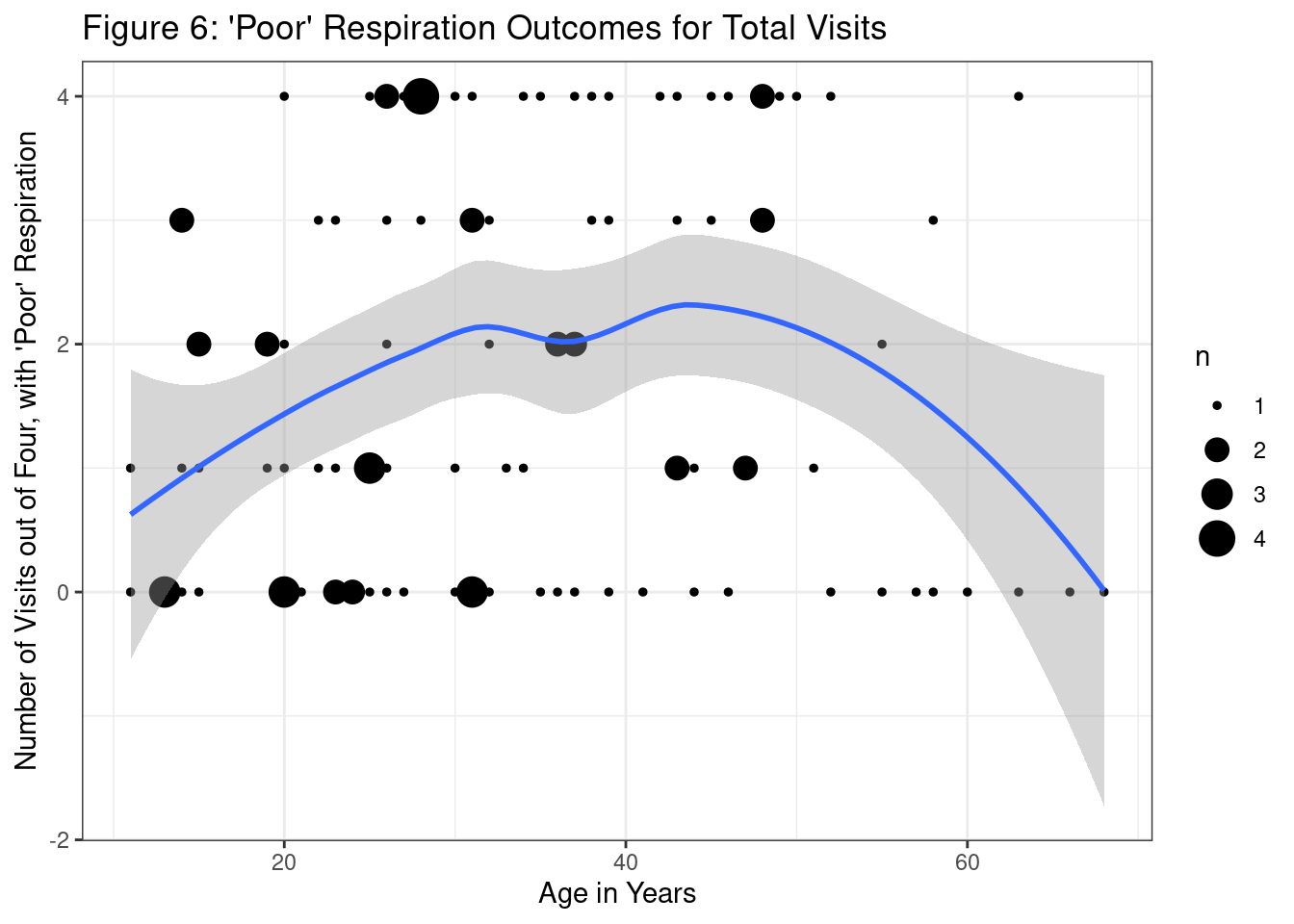
Agresti, Alan. 2015. Foundations of Linear and Generalized Linear Models. John Wiley & Sons.
Barnhart, Huiman X, and John M Williamson. 1998. “Goodness-of-Fit Tests for GEE Modeling with Binary Responses.” Biometrics, 720–29.
Burton, Lyle Gurrin, Paul, and Peter Sly. 1998.
“Extending the Simple Linear Regression Model to Account for Correlated Responses: An Introduction to Generalized Estimating Equations and Multi-Level Mixed Modelling.” Statistics in Medicine 17 (11): 1261–91.
https://doi.org/10.1002/(SICI)1097-0258(19980615)17:11<1261::AID-SIM846>3.0.CO;2-Z.
Crespi, Catherine M, Weng Kee Wong, and Shiraz I Mishra. 2009. “Using Second-Order Generalized Estimating Equations to Model Heterogeneous Intraclass Correlation in Cluster-Randomized Trials.” Statistics in Medicine 28 (5): 814–27.
D’Angelo, Gina M, Nicole A Lazar, William F Eddy, John C Morris, and Yvette I Sheline. 2011. “A Generalized Estimating Equations Approach for Resting-State Functional MRI Group Analysis.” In 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, 5064–67. IEEE.
Donald, Hedeker, and D Gibbons Robert. 2006. “Longitudinal Data Analysis.” John Wiley & Sons, Inc., Hoboken, NJ. DOI 10: 0470036486.
Garson, GD. 2013. “Generalized Linear Models/Generalized Estimating Equations. 2013 Ed.” Asheboro, NC: Statistical Associates Publishers.
Gayen, Atin, and M Ashok Kumar. 2018. “Generalized Estimating Equation for the Student-t Distributions.” In 2018 IEEE International Symposium on Information Theory (ISIT), 571–75. IEEE.
Halekoh, Ulrich, Søren Højsgaard, and Jun Yan. 2006. “The r Package Geepack for Generalized Estimating Equations.” Journal of Statistical Software 15/2: 1–11.
Hanley, James A, Abdissa Negassa, Michael D deB Edwardes, and Janet E Forrester. 2003. “Statistical Analysis of Correlated Data Using Generalized Estimating Equations: An Orientation.” American Journal of Epidemiology 157 (4): 364–75.
Hardin, J, and J Hilbe. 2013. “Generalized Estimating Equations (Second.).” New York: CRC Press.
Hin, Lin-Yee, and You-Gan Wang. 2009. “Working-Correlation-Structure Identification in Generalized Estimating Equations.” Statistics in Medicine 28 (4): 642–58.
Homish, Gregory G, Ellen P Edwards, Rina D Eiden, and Kenneth E Leonard. 2010. “Analyzing Family Data: A GEE Approach for Substance Use Researchers.” Addictive Behaviors 35 (6): 558–63.
Huang, Francis L. 2022.
“Analyzing Cross-Sectionally Clustered Data Using Generalized Estimating Equations.” Journal of Educational and Behavioral Statistics 47 (1): 101–25.
https://doi.org/10.3102/10769986211017480.
Hubbard, Alan E, Jennifer Ahern, Nancy L Fleischer, Mark Van der Laan, Sheri A Satariano, Nicholas Jewell, Tim Bruckner, and William A Satariano. 2010. “To GEE or Not to GEE: Comparing Population Average and Mixed Models for Estimating the Associations Between Neighborhood Risk Factors and Health.” Epidemiology, 467–74.
Huh, David, Brian P Flaherty, and Jane M Simoni. 2012. “Optimizing the Analysis of Adherence Interventions Using Logistic Generalized Estimating Equations.” AIDS and Behavior 16: 422–31.
Ito, Tsubasa, and Shonosuke Sugasawa. 2022. “Grouped Generalized Estimating Equations for Longitudinal Data Analysis.” Biometrics.
Koch, Gary G, Gregory J Carr, Ingrid A Amara, Maura E Stokes, and Thomas J Uryniak. 1990. “Categorical Data Analysis.” Statistical Methodology in the Pharmaceutical Sciences 13: 389–470.
Koper, Nicola, and Micheline Manseau. 2009. “Generalized Estimating Equations and Generalized Linear Mixed-Effects Models for Modelling Resource Selection.” Journal of Applied Ecology, 590–99.
Liang, Kung-Yee, and Scott L Zeger. 1986. “Longitudinal Data Analysis Using Generalized Linear Models.” Biometrika 73 (1): 13–22.
Lin, Meng-Lung, and Cheng-Hwang Perng. 2011. “The Impact of Terrain on NDVI Dynamics of Corn Field Using Generalized Estimating Equations and Time-Series MODIS Images.” In 2011 IEEE International Geoscience and Remote Sensing Symposium, 3342–45. IEEE.
Luo, Renwen, and Jianxin Pan. 2022. “Conditional Generalized Estimating Equations of Mean-Variance-Correlation for Clustered Data.” Computational Statistics & Data Analysis 168: 107386.
McCulloch, Charles E. 1996. “An Introduction to Generalized Linear Mixed Models.”
McCulloch, Searle, C. 2008.
Generalized, Linear, and Mixed Models, 2nd Edition. Wiley Series in Probability and Statistics. Wiley New Jersey.
https://www.wiley.com/en-us/Generalized%2C+Linear%2C+and+Mixed+Models%2C+2nd+Edition-p-9780470073711.
McDaniel, Lee S, Nicholas C Henderson, and Paul J Rathouz. 2013. “Fast Pure r Implementation of GEE: Application of the Matrix Package.” The R Journal 5 (1): 181.
Nam, Ju-Hyun, Myeong-Seob Lim, Hyun-Kyeong Choi, Jae-Yeop Kim, Sung-Kyeong Kim, Sung-Soo Oh, Sang-Baek Koh, and Hee-Tae Kang. 2017. “Factors Increasing the Risk for Psychosocial Stress Among Korean Adults Living in Rural Areas: Using Generalized Estimating Equations and Mixed Models.” Annals of Occupational and Environmental Medicine 29: 1–12.
Overall, John E, and Scott Tonidandel. 2004. “Robustness of Generalized Estimating Equation (GEE) Tests of Significance Against Misspecification of the Error Structure Model.” Biometrical Journal: Journal of Mathematical Methods in Biosciences 46 (2): 203–13.
Paik, Myunghee C. 1988. “Repeated Measurement Analysis for Nonnormal Data in Small Samples.” Communications in Statistics-Simulation and Computation 17 (4): 1155–71.
Pan, Wei. 2001. “Akaike’s Information Criterion in Generalized Estimating Equations.” Biometrics 57 (1): 120–25.
Paradis, Emmanuel, and Julien Claude. 2002. “Analysis of Comparative Data Using Generalized Estimating Equations.” Journal of Theoretical Biology 218 (2): 175–85.
Pekár, Stano, and Marek Brabec. 2018. “Generalized Estimating Equations: A Pragmatic and Flexible Approach to the Marginal GLM Modelling of Correlated Data in the Behavioural Sciences.” Ethology 124 (2): 86–93.
Salazar, Alejandro, Begoña Ojeda, Marı́a Dueñas, Fernando Fernández, and Inmaculada Failde. 2016. “Simple Generalized Estimating Equations (GEEs) and Weighted Generalized Estimating Equations (WGEEs) in Longitudinal Studies with Dropouts: Guidelines and Implementation in r.” Statistics in Medicine 35 (19): 3424–48.
Schwartz S, Barrett T. 2023.
“Encyclopedia of Quantitative Methods in r, Vol 5, 15-19: GEE.” 2023.
https://cehs-research.github.io/eBook_multilevel/gee-binary-outcome-respiratory-illness.html.
Seals, Samantha R, and Inmaculada B Aban. 2016. “Analysis of the 17-Segment Left Ventricle Model Using Generalized Estimating Equations.” Journal of Nuclear Cardiology. Springer.
Sepato, Sandra Moepeng. 2014. “Generalized Linear Mixed Model and Generalized Estimating Equation for Binary Longitudinal Data.” PhD thesis, University of Pretoria.
Shao, Sijing, Judith E Canner, Rebecca A Everett, Kidist Bekele-Maxwell, Alexis Kuerbis, Lyric Stephenson, Jennifer Menda, Jon Morgenstern, and HT Banks. 2023. “A Comparison of Mathematical and Statistical Modeling with Longitudinal Data: An Application to Ecological Momentary Assessment of Behavior Change in Individuals with Alcohol Use Disorder.” Bulletin of Mathematical Biology 85 (1): 5.
Statistics, Penn State Department of. 2023.
“Analysis of Discrete Data, 12: Advanced Topics II.” 2023.
https://online.stat.psu.edu/stat504/lesson/12/12.2.
Touloumis, Anestis. 2014. “R Package Multgee: A Generalized Estimating Equations Solver for Multinomial Responses.” arXiv Preprint arXiv:1410.5232.
V, Carey. 2022.
“Gee: Generalized Estimation Equation Solver.” 2022.
https://cran.r-project.org/package=gee.
Wang, Lan, Jianhui Zhou, and Annie Qu. 2012. “Penalized Generalized Estimating Equations for High-Dimensional Longitudinal Data Analysis.” Biometrics 68 (2): 353–60.
Wang, Ming. 2014. “Generalized Estimating Equations in Longitudinal Data Analysis: A Review and Recent Developments.” Advances in Statistics 2014.
Warton, David I. 2011. “Regularized Sandwich Estimators for Analysis of High-Dimensional Data Using Generalized Estimating Equations.” Biometrics 67 (1): 116–23.
Yan, Jun. 2002. “Geepack: Yet Another Package for Generalized Estimating Equations.” R-News 2/3: 12–14.
Yan, Jun, and Jason P. Fine. 2004. “Estimating Equations for Association Structures.” Statistics in Medicine 23: 859–80.
Zeileis, Achim, Thomas Lumley, Susanne Berger, Nathaniel Graham, and Maintainer Achim Zeileis. 2019. “Package ‘Sandwich’.” R Package Version, 2–5.