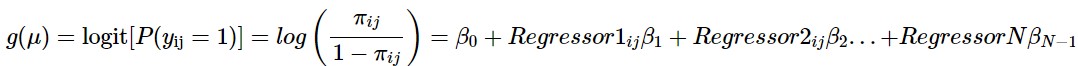Call:
geeglm(formula = outcome ~ center + treat + age + baseline +
sex, family = binomial(), data = respiratory, id = id, corstr = "independence")
Coefficients:
Estimate Std.err Wald Pr(>|W|)
(Intercept) -0.10346 0.88213 0.014 0.906634
center 0.64949 0.35322 3.381 0.065952 .
treatP -1.26536 0.34668 13.322 0.000262 ***
age -0.01876 0.01296 2.093 0.147974
baseline 1.84572 0.34598 28.460 9.57e-08 ***
sexM -0.13678 0.44025 0.097 0.756035
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation structure = independence
Estimated Scale Parameters:
Estimate Std.err
(Intercept) 1.002 0.1842
Number of clusters: 111 Maximum cluster size: 4 Generalized Estimating Equations
2023-07-31
Introduction - What is GEE?
Generalized Estimating Equations (GEE) is a statistical methodology that expands upon generalized linear models.
The method was first introduced by Liang and Zeger in a paper in 1986. (Liang and Zeger 1986).
Open to the possibility of correlation within the data (contrary to most methodologies that assume independence of observations)
Introduction - When to use GEE?
Usually longitudinal data or cluster data (epidemiology, social sciences, clinical trials, etc.). (S. McCulloch C. 2008)
GEE does not require a specified distribution
GEE does require a specified relationship between mean and variance
The method uses a quasilikelihood approach and yields population averaged estimates of parameters. (M. Wang 2014)
Methods
Estimating Equations - Quasilikelihood approach to estimating parameters
Iterative Algorithm - Using IRLS to iterate estimates of \(a\) to get new estimates for \(\beta\) until convergence
Data Structure - Used for analyzing correlated, such as longitudinal or panel data
Model Specification - Extension of GLM, models can be for countinuous, binary or count outcomes
Correlation Structures - Flexible working correlation structures available based on data
Methods - Estimating Equations
In Wang’s review of GEE method, a marginal model specifying a relationship between \(\mu_i\) and \(X_{ij}\) is given by the equation \(g(\mu_{ij})=X'_{ij} \beta\), where \(g\) is the link function and \(\beta\) is a vector of regression coefficients and then shows the mathematical calculations to get to the equation
\(U(\beta) = \sum_{i=1}^K {D_i}^{\prime} {V_i}^{-1}(Y_i-\mu_i)=0\)
that when solved will give estimates of \(\beta_i\) used in the generated models.(M. Wang 2014)
Methods - Estimating Equations, continued
In this equation \(D_i=\partial\mu_i/\partial\beta^{\prime}\) and \({V_i}^{-1}\) is the inverse of the variance-covariance matrix for \(Y_i\), \(V_i = \phi{A_i}^{1/2}R_i(a){A_i}^{1/2}\) , \(A_i\) is a diagonal matrix with elements that are known variance functions of \(\mu_{ij}\) times a scaling parameter \(\phi\), and \(R_i(a)\) is the correlation structure. (M. Wang 2014)
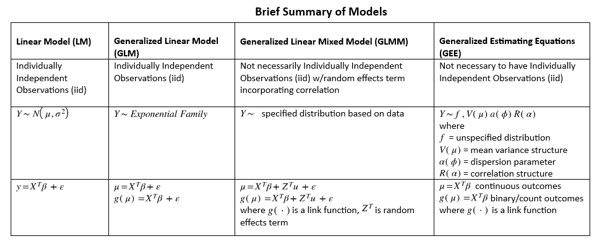
Methods - Model Specification
Methods - Correlation Structures in ‘geepack’
- Independent - Assumption that all individual observations are independent
- Exchangeable - Assumption there is equality of time between observations
- Autoregressive (AR1) - Assumption that there are more than one set of equality of times between observations
- Unstructured - Most general and takes into account all possible times possible between observations
Methods - Correlation Structures
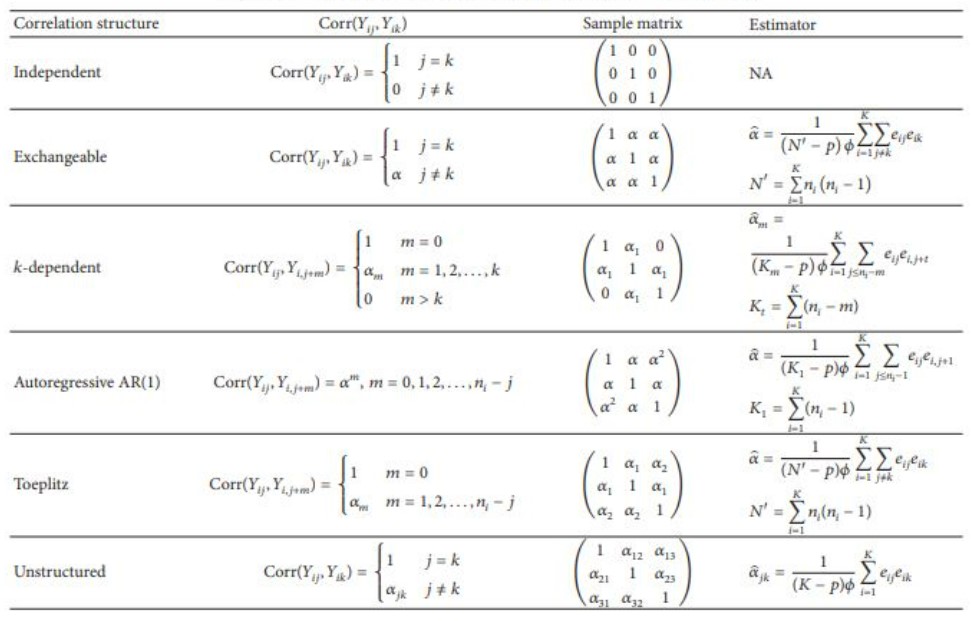
Figure 2: Correlation Structures Comparison Table (M. Wang 2014)
Analysis and Results - Dataset
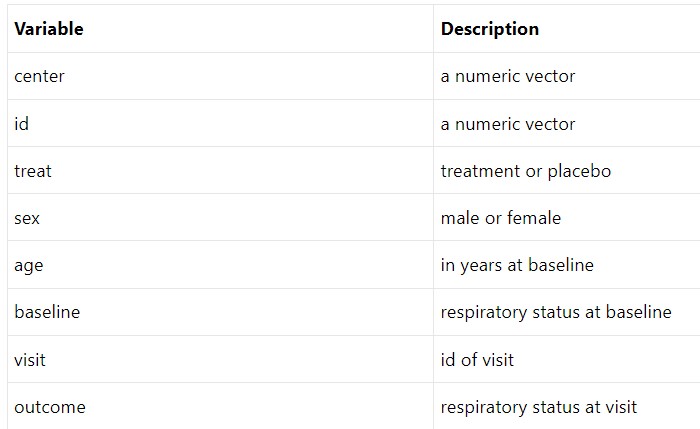
Figure 3: Respiratory Variable Summary (Halekoh, Højsgaard, and Yan 2006)
Analysis and Results: Sample Data Plot
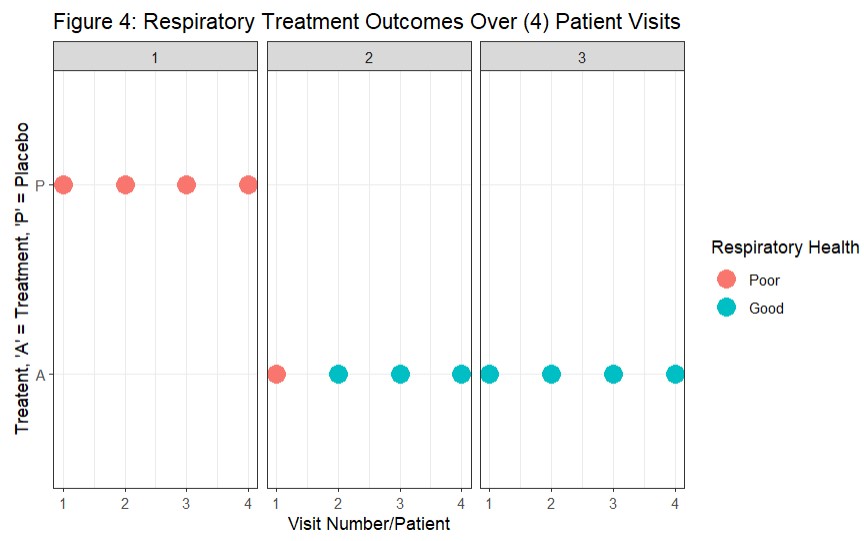
Figure 4: Respiratory Treatment Outcomes Over (4) Patient Visits (Halekoh, Højsgaard, and Yan 2006)
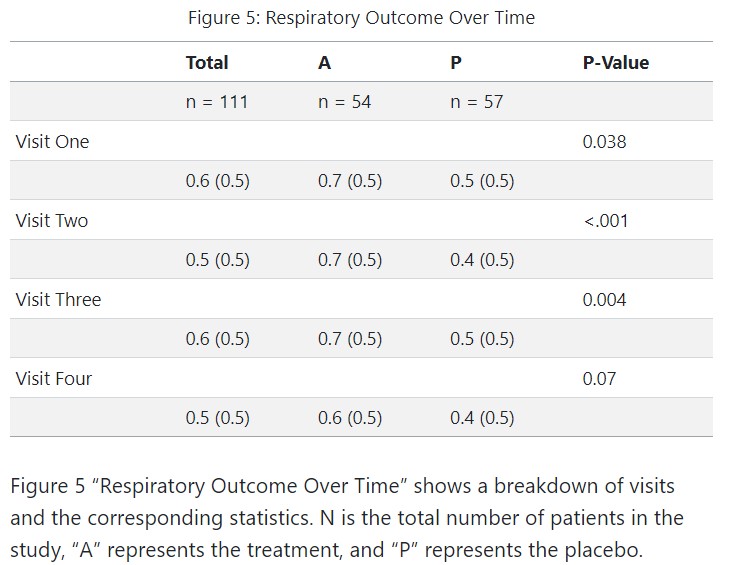
Analysis and Results: Outcomes Over Time
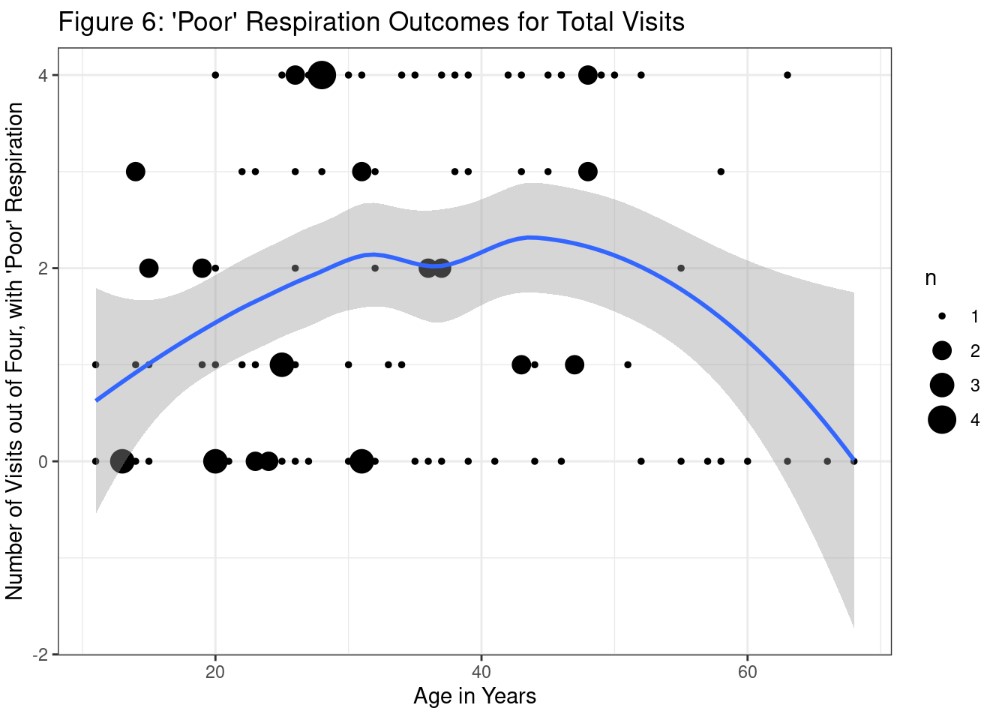
Analysis and Results: ‘Poor’ Outcomes for Total Visits
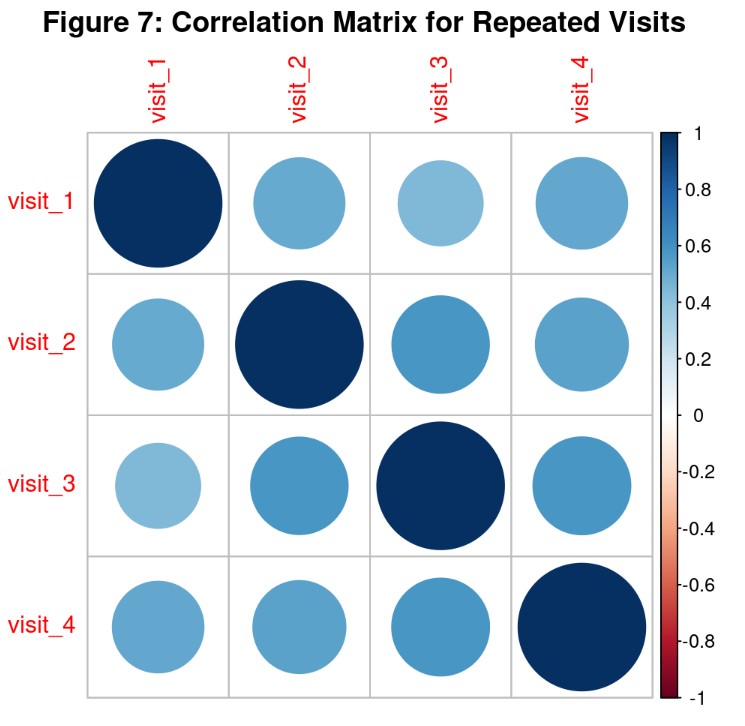
Analysis and Results: Correlation Matrix For Repeated Visits
Statistical Modeling
GEE Model(s)
Model Assessment
QIC (Quasi Information Criterion) for Model
Inference
Statistical Modeling: GG w/Independent Correlation Structure
Statistical Modeling: GG w/Exchangeable Correlation Structure
Call:
geeglm(formula = outcome ~ center + treat + age + baseline +
sex, family = binomial(), data = respiratory, id = id, corstr = "exchangeable")
Coefficients:
Estimate Std.err Wald Pr(>|W|)
(Intercept) -0.1035 0.8821 0.01 0.90663
center 0.6495 0.3532 3.38 0.06595 .
treatP -1.2654 0.3467 13.32 0.00026 ***
age -0.0188 0.0130 2.09 0.14797
baseline 1.8457 0.3460 28.46 9.6e-08 ***
sexM -0.1368 0.4402 0.10 0.75604
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation structure = exchangeable
Estimated Scale Parameters:
Estimate Std.err
(Intercept) 1 0.184
Link = identity
Estimated Correlation Parameters:
Estimate Std.err
alpha 0.329 0.0838
Number of clusters: 111 Maximum cluster size: 4 Statistical Modeling: GG w/Autoregressive Correlation Structure
Call:
geeglm(formula = outcome ~ center + treat + age + baseline +
sex, family = binomial(), data = respiratory, id = id, corstr = "ar1")
Coefficients:
Estimate Std.err Wald Pr(>|W|)
(Intercept) -0.3231 0.8716 0.14 0.71083
center 0.7297 0.3525 4.29 0.03843 *
treatP -1.1914 0.3483 11.70 0.00062 ***
age -0.0174 0.0129 1.83 0.17647
baseline 1.8709 0.3466 29.14 6.7e-08 ***
sexM -0.1346 0.4477 0.09 0.76365
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation structure = ar1
Estimated Scale Parameters:
Estimate Std.err
(Intercept) 1.01 0.19
Link = identity
Estimated Correlation Parameters:
Estimate Std.err
alpha 0.457 0.0896
Number of clusters: 111 Maximum cluster size: 4 Statistical Modeling: GG w/Unstructured Correlation Structure
Call:
geeglm(formula = outcome ~ center + treat + age + baseline +
sex, family = binomial(), data = respiratory, id = id, corstr = "unstructured")
Coefficients:
Estimate Std.err Wald Pr(>|W|)
(Intercept) -0.1826 0.8685 0.04 0.83351
center 0.6555 0.3513 3.48 0.06203 .
treatP -1.2455 0.3455 12.99 0.00031 ***
age -0.0176 0.0129 1.87 0.17147
baseline 1.8944 0.3441 30.31 3.7e-08 ***
sexM -0.1144 0.4407 0.07 0.79528
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation structure = unstructured
Estimated Scale Parameters:
Estimate Std.err
(Intercept) 1.01 0.194
Link = identity
Estimated Correlation Parameters:
Estimate Std.err
alpha.1:2 0.321 0.118
alpha.1:3 0.206 0.101
alpha.1:4 0.284 0.110
alpha.2:3 0.424 0.141
alpha.2:4 0.343 0.119
alpha.3:4 0.380 0.133
Number of clusters: 111 Maximum cluster size: 4 alpha.1:2 alpha.1:3 alpha.1:4 alpha.2:3 alpha.2:4 alpha.3:4
0.321 0.206 0.284 0.424 0.343 0.380 Statistical Modeling: Model Assessment
The following formula is the logistic regression model unique to the respiratory data set. This formula was translated into the four different models above with a different correlation structure specified per model.
\[ \log\left(\dfrac{\pi_{ij}}{1-\pi_{ij}}\right)=\beta_0+Center_{ij}\beta_1+Treat_i\beta_2+Age_i\beta_3+Sex_i\beta_4+Baseline_i\beta_5 \] (Halekoh, Højsgaard, and Yan 2006)
Statistical Modeling: QIC for Model Selection
QIC QICu Quasi Lik CIC params QICC
gee_independence 513 499 -244 12.5 6 514
gee_exchangeable 513 499 -244 12.5 6 515
gee_unstructured 512 500 -244 12.4 6 520
gee_ar1 513 500 -244 12.7 6 516Statistical Modeling: Inference
GEE provides robust standard errors for the estimated regression parameters, taking into account the correlation structure. When comparing multiple different techniques, it’s important to consider the correlation between observations in order to perform accurate statistical inference of the model’s output. (Seals and Aban 2016) Hypothesis tests and confidence intervals can be constructed to assess the statistical significance of the variables and make inferences about the population-level effects. (Shao et al. 2023)
Conclusion
Use GEE in cases when assumptions for linear regression models and generalized linear models do not hold (non-normal data, dependence in observations, longitudinal or panel data, clustered data, correlated data, etc.)
Use GEE when there are large amounts of higher dimensional data (finding the likelihood functions is very difficult computationally)
GEE provides an easy alternative and gives stable, consistent estimates for parameters without the need for the possibly intense likelihood calculations.
GEE can be used for continuous, binary or count responses.